Better Science with Spooky Tech
By Josi Livingston • October 31, 2024

In countless conversations about AI in the life sciences, a recurring theme emerges: the pressing need for vast, clean, "AI-ready" data. People often envision AI as massive prediction engines like AlphaFold or other drug discovery tools that rely on enormous databases of protein sequences and omics data. While these are undeniably important applications, it's unfortunate that other, less glamorous (yet equally critical) uses receive less attention in pharmaceutical sciences. One such area is synonym management and ontology harmonisation, where AI's semantic awareness holds immense potential.
To explain why—and to illustrate the frustration of dealing with systems that don't understand semantics (and since it's Halloween)—let me share a story about a Halloween-themed 8-bit text parser adventure game from the '90s called Hugo's House of Horrors.
If you're too young (or possibly too cool) to remember, this game casts you as Hugo, whose girlfriend Penelope has been kidnapped following an ill-advised babysitting job at an obviously haunted house. Your mission is to explore the house and rescue her by directing Hugo with arrow keys and simple commands like "open door" or "inspect mask." Along the way, you're met with various spooky puzzles and nefarious monsters that frustrate your efforts. More often than not, however, the most difficult task was overcoming the game's restrictive parser engine.

Your first puzzle is to unlock the front door using a key hidden somewhere. Anyone who has ever played any video game ever can probably tell right away that the key is hidden in that bright yellow pumpkin out front.
When I first played this game as a young child, I tried every command I could think of to get this key, but nothing worked. "Open pumpkin," "get pumpkin," "look pumpkin," "inspect pumpkin," "smash the stupid pumpkin"—I tried them all. Hours passed banging my head against this problem, until my older brother came around and pointed out (rather smugly) that I had a typo in my command. I had misspelled "pumpkin" as "pumkin." Sure enough, once I spelled it correctly, I had the key, I was in the mansion, and I developed a chip on my shoulder about computer interfaces that would persist for three decades.
Because here's the thing: did I send the wrong command to a rudimentary parser of limited capability? Yes. But was my logic faulty? No. Semantically speaking, I was correct even if orthographically speaking I was wrong—which seems like a harsh standard to hold when poor Penelope's life is on the line.
I think about this all the time when I think about modern AI, because nomenclature problems are everywhere in the life sciences but these new ChatGPT/Claude/LLM tools are incredible at understanding semantics, even when the source text is imprecise. The maths behind how it works is fascinating but complex. If you're curious to dig into it I really like 3Blue1Brown's explanation of the mechanics in this video, which I highly recommend.
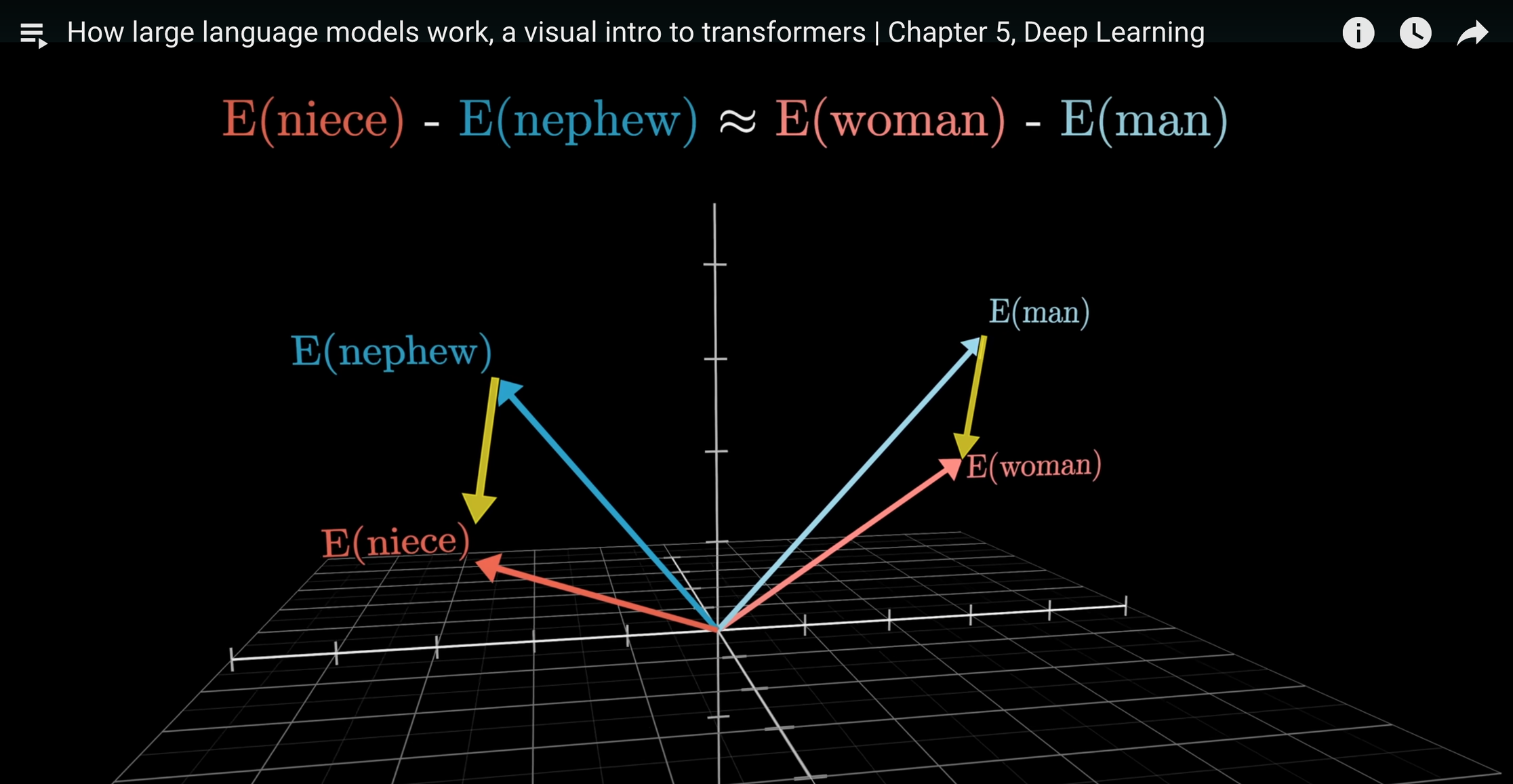
You should really watch the whole thing, but my favourite part is around minute 15. There, he explains that if you represent all words as numerical vectors in a multidimensional space (which is what LLMs are doing—they're called "embeddings"), you can perform arithmetic on them to uncover surprising results. He gives the classic example about how the difference between the vectors of "man" and "woman" is approximately the same as the difference between "king" and "queen," or "nephew" and "niece." This implies that the machine has learned, through no explicit instruction, that directionality in this space has semantic meaning. That is, one of the directions represents "femaleness," another "bigness," a third "yellowness," etc.
These aren't hard-and-fast associations because the algorithm invents them independently during pre-training. But as a fun exercise, it turns out that you can explicitly look at these dimensions and infer an intrinsic meaning about them after the fact. My code for inspecting token embeddings in a model, performed in Google Colab
For example, Meta's Llama-2-7B model represents each of its words (technically, "tokens," but we don't need to get into all that right now) with a vector of 4,096 numbers. This means that the embedding space has 4,096 separate dimensions and therefore 4,096 different types of semantic meaning.
Suppose we look at Dimension 13 (since it's Halloween and 13 is a spooky number). The top-scoring positive words are "generalized," "backup," "arrival," and "quadratic," while the top-scoring negative words are "professional," "wanting," "fiddle," and "preference." One possible way to interpret this is that Dimension 13 captures the duality between the safety of a multi-purpose math career (say, accounting) and the hidden longing of someone wishing to explore the adventurous life of a musician. It's fun to think that all words contain some tiny essence of this linear scale.
This example is whimsical, but the implications are profound. Because of the way LLMs work, a word isn't meaningful because of its dictionary definition or properties in a predefined database. It's meaningful because it has been shaped by the meanings of words around it. That's why if you talk to an LLM about Cyclohexa-1,3,5-triene—a chemical with CAS no. 71-43-2—or benzene, it understands that these three things are equivalent because they are discussed in equivalent ways in its original training material.
I mention all this because, in the sciences generally and in the medical/pharmaceutical sciences especially, we have a real synonym and ontology problem that gets in the way of progress. I encountered this early in my career as a consultant when I had a client who wanted to perform a human health risk assessment for hazardous waste incineration. The chemical nomenclature example I just gave is particularly relevant, as performing such an analysis requires sifting through dozens of databases, each of which indexes chemicals differently. This challenge is compounded by the industry's frequent use of trade names or historical terms for chemicals—practices that range from archaic at best to deliberately misleading at worst. At the time, I worked very hard to ensure that I didn't have any synonym issues in my report, but it disturbs me to think that we are one "pumkin" v. "pumpkin" error away from causing harm.
This is why common tools like LIMS and ELNs exist for biotechnology. Having worked for vendors in this space for well over a decade, I've seen firsthand how much effort and treasure clients put toward the simple task of nomenclature harmonisation, for fear of this very problem. Months are spent negotiating between teams on how to name things, which dropdown options will be valid, how to enforce data validation during data entry in an electronic notebook, how to put data into a warehouse and pull it back out again, and on and on and on. It is not only expensive but fundamentally flawed because it requires pre-anticipation of every contingency in a domain that is, by its nature, dynamic and emergent. People may bemoan "Excel Hell," but we have found ourselves in "Platform Purgatory".
This is why modern AI excites me so much right now. For decades, our tools have become more complex and powerful, but we've had to bend our thinking and processes to conform to their paradigms. We are Hugo trying to save Penelope through the power of proper syntax. But for the first time in a long time, technology is meeting us at our level. To put it another way, we have spent decades as humans becoming more machine-like, but now machines are becoming more human. This phenomenon creeps out many, but Halloween is the perfect day to find joy in something a little spooky.
While some worry about AI's readiness to handle complex scientific terminology, continuous training on specialised datasets enhances its precision and reliability. That's why in my last article I balked at the notion of "Garbage In, Garbage Out". Now more than ever, the value you can get from unstructured information, like historic notebooks or disparate laboratory logs, is boundless.
One example application could be this:
Use bulk text from your laboratory workflows to fine-tune a base LLM with specific terminology. Use that LLM to suggest a harmonised, use-case specific ontology. Align existing structured data to that ontology. Combined the structured data and use it to train a larger, predictive AI engine.
I've been working on a project the past few weeks demonstrating how to do parts of this, specifically, I'm training a base LLM on raw Alzheimer's disease journal articles to see how it impacts chat efficacy. I'll be wrapping that up shortly, but in the meantime, if you'd like help understanding how you can incorporate AI into your workflows today, drop me a line.